
|
Joshua Buckner, Xiarong Liu, Arghya Chakravorty, Yujin Wu, Luis Cervantes, Thanh Lai, Charles L. Brooks III: Journal of Chemical Theory and Computation 2023 19 (12), 3752-3762 |

|
Heng Wu, He Huang, Carol B. Post: J. Chem. Phys. (2020) 153 175101 |

|
Simone Conti, Kevin J. Kaczorowski, Ge Song, Katelyn Porter, Raiees Andrabi, Dennis R. Burton, Arup K. Chakraborty, Martin Karplus: PNAS (2021) 118, e2018338118 |

|
Wonmuk Hwang, Robert J. Mallis, Matthew J. Lang, Ellis L. Reinherz: PNAS (2020) 117, 21336-21345 |

|
Xinqiang Ding, Yujin Wu, Yanming Wang, Jonah Z. Vilseck, Charles L. Brooks III: JCTC (2020), 16, 3910-3919 |

|
Yalun Yu, Andreas Krämer, Richard M. Venable, Andrew C. Simmonett, Alexander D. MacKerell Jr., Jeffrey Klauda, Richard W. Pastor, Bernard R. Brooks: JCTC (2021), 17, 1562-1580 |
|
E. Prabhu Raman, Thomas J. Paul, Ryan L. Hayes, Charles L. Brooks III: JCTC (2020), 16, 7895-7914 |
|
Hedieh Torabifard, Afra Panahi, Charles L. Brooks III: PNAS (2020) 117, e1913385117 |
|
Yandong Huang, Wei Chen, David L. Dotson, Oliver Beckstein, Jana Shen: Nature Communications (2016), 7, 12940 |
|
Jing Huang, Sarah Rauscher, Grzegorz Nawrocki, Ting Ran, Michael Feig, Bert L. de Groot, Helmut Grubmüller, Alexander D. MacKerell Jr.: Nature Methods (2016), 14, 71-73
|
|
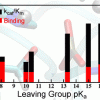
Daniel Roston, Qiang Cui: J. Am. Chem. Soc. (2016) 138, 11946-11957 |
|
Ning Ma, Arjan van der Vaart: J. Am. Chem. Soc. (2016) 138, 9951-9958 |
|
Evan J. Arthur, Charles L. Brooks: J. Comp. Chem. (2016) 37, 2171-2180 |

|
Isseki Yu, Takaharu Mori, Tadashi Ando, Ryuhei Harada, Jaewoon Jung, Yuji Sugita, Michael Feig: eLife (2016) 5, e19274 |
|
You Xu, Alexander D. MacKerell Jr., Lennart Nilsson: Bioorganic & Medicinal Chemistry (2016) 24, 4826-4834 |

|
Yilin Meng, Diwakar Shukla, Vijay S. Pande, Benoit Roux: PNAS (2016) 113, 9193-9198 |

|
Yen-Lin Lin, Yilin Meng, Lei Huang, Benoit Roux: J. Am. Chem. Soc. (2014) 136, 14753-14762 |

|
Xiancheng Zeng, Suchetana Mukhopadhyay, Charles L. Brooks III: Proc. Natl. Acad. Sci. USA (2015) 112, 2034-2039 |
|
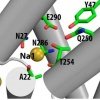
Sebastian Stolzenberg, Matthias Quick, Chunfeng Zhao, Kamil Gotfryd, George Khelashvili, Ulrik Gether, Claus J. Loland, Jonathan A. Javitch, Sergei Noskov, Harel Weinstein, Lei Shi: J. Biol. Chem. (2015) 290, 13992-14003 |
|
Rodney E. Versace, Themis Lazaridis: J. Phys. Chem. B (2015) 119, 8037-8047 |



















